Software
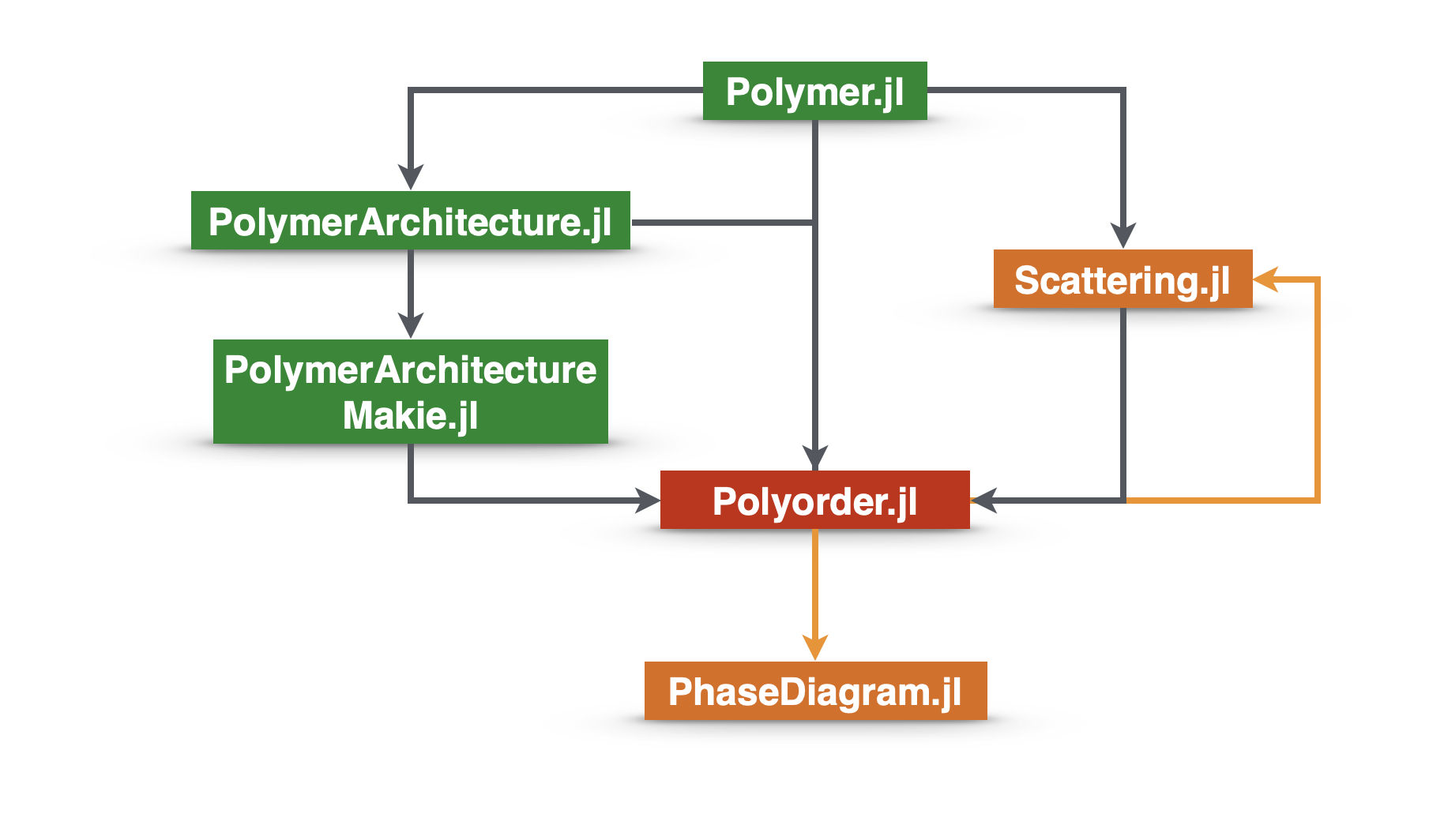
Some listed software are open sourced. More will be open sourced as they become mature. Please check our github.com account for the latest updates.
Scientific Computing
Polyorder.jl
A pure Julia implementation of polymer self-consistent field theory
Features
- Support arbitrary noncyclic chain architectures
- Modern acceleration algorithms (Anderson mixing, GMRES)
- Support 1D, 2D, and 3D unit cells
- RPA analysis support
Links
Scattering.jl
Julia package for computing scattering and diffraction of nanoparticles
Features
- Form factor calculations
- Structure factor calculations
- Translation and rotation operations
Links
PhaseDiagram.jl
Julia package for computing phase diagrams of block copolymers
Features
- Microphase separation diagrams
- Macrophase separation diagrams
Polymer.jl
Common interface for describing polymer systems
Installation
1
2
julia> ]
pkg> add Polymer
Links
PolymerArchitecture.jl
Graph representation of polymer architectures
Features
- Automatic identification of equivalent sub-architectures
- Integration with Polymer.jl
CubicHermiteSpline.jl
Cubic Hermite spline interpolation for 1D and 2D data
Features
- Univariate and bivariate interpolation
- Gradient calculations
Installation
1
2
julia> ]
pkg> add CubicHermiteSpline
Links
Utility
MakiePublication.jl
Publication quality figures using Makie.jl
Features
- Custom themes for major publishers
- 15 color palettes
- Hollow markers support
Links
PolymerArchitectureMakie.jl
Visualization of polymer architectures using GraphMakie.jl
Features
- Integration with PolymerArchitecture.jl
- Customizable visual styles
mpltex
Python package for creating publication-quality plots
Features
- ACS style plots
- Presentation and web styles
- Tableau color scheme
Links
Legacy
PolyOrder
C++ library for polymer self-consistent field theory (deprecated)
Features
- Non-orthogonal unit cell support
- Weakly charged polymers support
cheb++
C++ library for Chebyshev polynomials
Features
- Chebyshev differential matrix
- Clenshaw-Curtis quadrature
Links
scftpy
Python package for polymer SCFT calculations
Features
- Confined polymer simulations
- Polymer brush simulations
Links
chebpy
Python package for spectral methods of PDEs
Features
- Chebyshev series construction
- Fast Chebyshev transform
Links
PyDiagram
Python package for generating phase diagrams
Features
- Polyorder and PolyFTS file support
- Free energy analysis
- Phase boundary plotting
gyroid
Python package for symmetry adapted basis functions
Features
- 1D, 2D and 3D symmetry groups
- Structure renderer
Links
NGPy
Web application for nucleation and growth simulations
Features
- Online simulation and analysis
- Web framework for custom applications